-
- Posts: 7
- Joined: Tue Mar 15, 2005 7:24 pm
The chromatogram below is from a standard injection, chloroform concentration in DMF = 3.7 ng/mL. Chloroform r.t. = 2.24 min.
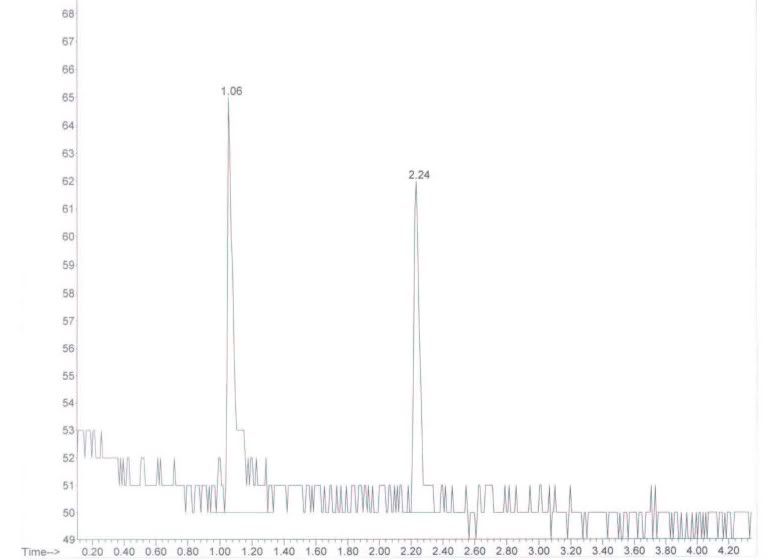
My first question is one concerning the baseline noise. Is this pattern typical when performing trace analysis w/ SIM? Can anyone suggest ways to minimize the effect of such noise on my integration? (In case you haven't guessed, I'm fairly new to SIM!)
Background: The GC method I'm running was validated for GC-FID w/ a LOQ of 95 ppm. W/ MSD, I'm getting %RSD < 3% for 6 replicate injections of 7.5 ng/mL standard and linearity is good (R^2 = 0.998 over 3.7 - 11.2 ng/mL range).
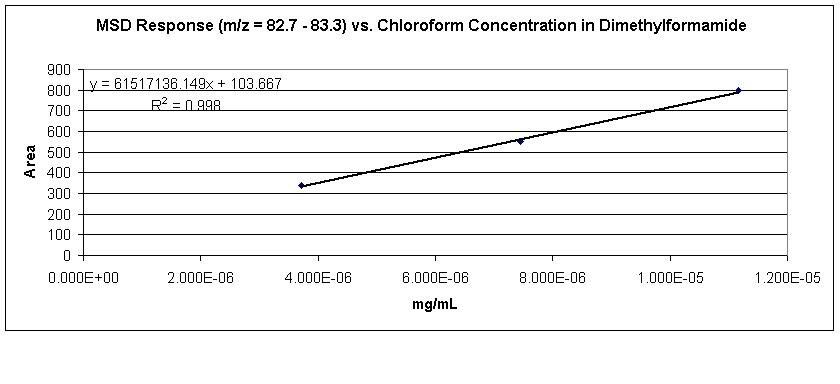
The problem I face is that my sample solutions appear to contain less than my lowest standard concentration. (Sample peak areas range from 120 to 218). S/N (pk-pk) = 11.5 for my 3.7 ng/mL standard, but only 1.7 for my least-concentrated sample.
Am I simply pushing the LOD for this system, or is there a way to further increase the sensitivity?
Any thoughts would be greatly appreciated. Thanks in advance!
FYI:
2 uL injection
20:1 split (Helium, split flow = 40 mL/min)
Column: Restek RTX-5 30m x 0.32 x 1 um film
